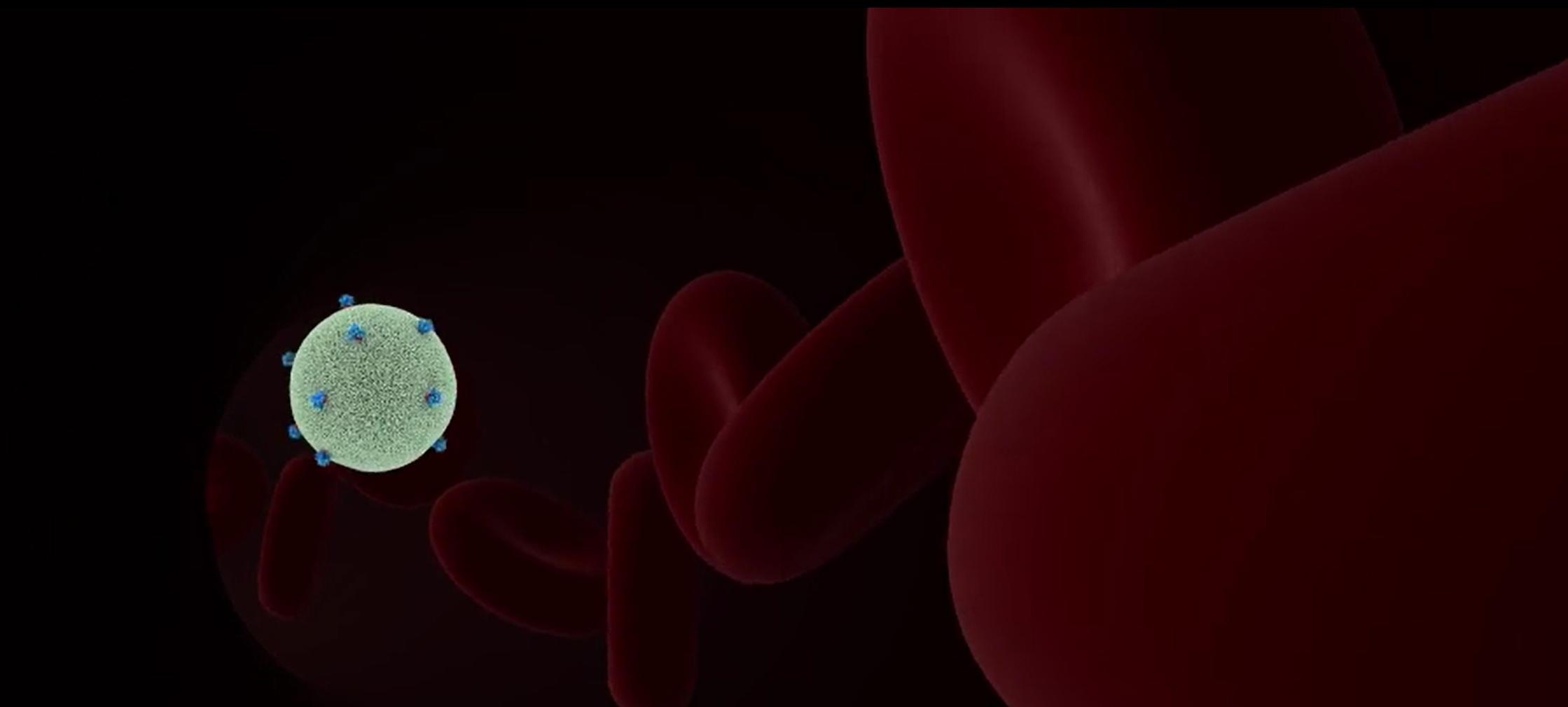
Innate Immunity
The innate immune response is our body’s first line of defense against infections by bacteria or viruses. During HIV infection, cells can recognize specific signals given by an invading virus and trigger a signaling cascade that alerts the rest of the body to the infection. Unfortunately, HIV has developed ways to evade and hide from our innate immune system, making it difficult to fight against the infection. To assist in this battle, researchers have been working hard to better understand how to activate our innate immune system in response to HIV. The following animations depict research based scenarios in which innate immune sensors are able to detect HIV and trigger an innate immune response.
cGAS-STING Pathway
Cyclic GMP-AMP synthase or cGAS plays an important role in detecting foreign double-stranded DNA in the cytoplasm. When cGAS detects and binds DNA, it produces a secondary messenger called, cGAMP, which then goes on to activate the adaptor protein STING. From there, a signaling cascade occurs leading to the activation of the transcription factor IRF3. This ultimately leads the release of interferon related proteins that then alert neighboring cells to activate their own interferon response, thereby creating a protective cellular environment that prevents the spread of HIV. In order to activate the cGAS-STING pathway, researchers have been developing methods to crack open the HIV-1 capsid while in the cytoplasm and before integration can occur. By using small molecules such as Lenacapavir, the HIV-1 DNA can become exposed to the cytoplasm thereby allowing cGAS to detect and bind the intruder.
MDA5 Activation
MDA5 is a cytosolic viral RNA sensor that can bind viral double-stranded RNA through its c-shaped helicase domain. Subsequent activation of MDA5 has been shown to lead to a signaling cascade that results in increased levels of pro-inflammatory cytokines. During the life cycle of HIV-1, unspliced RNA produced by the HIV-1 provirus is exported from the nucleus to the cytoplasm. This unspliced HIV-1 RNA has been shown to activate a type 1 interferon response, but for many years the innate immune receptor that detects unspliced HIV-1 RNA was unknown. Fortunately, new research suggests that MDA5 plays a key role in this detection and may contribute to the inflammation people living with HIV-1 experience.
Animation created by Rachel Torrez in collaboration with Dr. Jarrod Johnson’s lab at the University of Utah
-
(1) Christensen, D. E.; Ganser-Pornillos, B. K.; Johnson, J. S.; Pornillos, O.; Sundquist, W. I. Reconstitution and Visualization of HIV-1 Capsid-Dependent Replication and Integration in Vitro. Science 2020, 370 (6513), eabc8420. https://doi.org/10.1126/science.abc8420.
(2) Zhang, X.; Wu, J.; Du, F.; Xu, H.; Sun, L.; Chen, Z.; Brautigam, C. A.; Zhang, X.; Chen, Z. J. The Cytosolic DNA Sensor cGAS Forms an Oligomeric Complex with DNA and Undergoes Switch-like Conformational Changes in the Activation Loop. Cell Reports 2014, 6 (3), 421–430. https://doi.org/10.1016/j.celrep.2014.01.003. (PDB: 4O6A)
(3) Shang, G.; Zhang, C.; Chen, Z. J.; Bai, X.; Zhang, X. Cryo-EM Structures of STING Reveal Its Mechanism of Activation by Cyclic GMP–AMP. Nature 2019, 567 (7748), 389–393. https://doi.org/10.1038/s41586-019-0998-5. (PDB: 6NT6, 6NT7)
(4) Sumner, R. P.; Harrison, L.; Touizer, E.; Peacock, T. P.; Spencer, M.; Zuliani-Alvarez, L.; Towers, G. J. Disrupting HIV-1 Capsid Formation Causes cGAS Sensing of Viral DNA. The EMBO Journal 2020, 39 (20), e103958. https://doi.org/10.15252/embj.2019103958.
(5) Altfeld, M.; Gale Jr, M. Innate Immunity against HIV-1 Infection. Nat Immunol 2015, 16 (6), 554–562. https://doi.org/10.1038/ni.3157.
(6) Zhang, C.; Shang, G.; Gui, X.; Zhang, X.; Bai, X.; Chen, Z. J. Structural Basis of STING Binding with and Phosphorylation by TBK1. Nature 2019, 567 (7748), 394–398. https://doi.org/10.1038/s41586-019-1000-2. (PDB: 6NT9)
(7) Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; Žídek, A.; Green, T.; Tunyasuvunakool, K.; Petersen, S.; Jumper, J.; Clancy, E.; Green, R.; Vora, A.; Lutfi, M.; Figurnov, M.; Cowie, A.; Hobbs, N.; Kohli, P.; Kleywegt, G.; Birney, E.; Hassabis, D.; Velankar, S. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Research 2022, 50 (D1), D439–D444. https://doi.org/10.1093/nar/gkab1061.
(8) Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; Bridgland, A.; Meyer, C.; Kohl, S. A. A.; Ballard, A. J.; Cowie, A.; Romera-Paredes, B.; Nikolov, S.; Jain, R.; Adler, J.; Back, T.; Petersen, S.; Reiman, D.; Clancy, E.; Zielinski, M.; Steinegger, M.; Pacholska, M.; Berghammer, T.; Bodenstein, S.; Silver, D.; Vinyals, O.; Senior, A. W.; Kavukcuoglu, K.; Kohli, P.; Hassabis, D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596 (7873), 583–589. https://doi.org/10.1038/s41586-021-03819-2.
Animation created by Rachel Torrez in collaboration with Dr. Jeremy Luban’s lab at the University of Massachusetts Medical School
-
(1) Guney, M. H.; Nagalekshmi, K.; McCauley, S. M.; Carbone, C.; Aydemir, O.; Luban, J. IFIH1 (MDA5) Is Required for Innate Immune Detection of Intron-Containing RNA Expressed from the HIV-1 Provirus. Proceedings of the National Academy of Sciences 2024, 121 (29), e2404349121. https://doi.org/10.1073/pnas.2404349121.
(2) Jayaraman, B.; Crosby, D. C.; Homer, C.; Ribeiro, I.; Mavor, D.; Frankel, A. D. RNA-Directed Remodeling of the HIV-1 Protein Rev Orchestrates Assembly of the Rev–Rev Response Element Complex. eLife 2014, 3, e04120. https://doi.org/10.7554/eLife.04120. (PDB: 4PMI)
(3) Güttler, T.; Madl, T.; Neumann, P.; Deichsel, D.; Corsini, L.; Monecke, T.; Ficner, R.; Sattler, M.; Görlich, D. NES Consensus Redefined by Structures of PKI-Type and Rev-Type Nuclear Export Signals Bound to CRM1. Nat Struct Mol Biol 2010, 17 (11), 1367–1376. https://doi.org/10.1038/nsmb.1931. (PDB: 3NC0)
(4) Wies, E.; Wang, M. K.; Maharaj, N. P.; Chen, K.; Zhou, S.; Finberg, R. W.; Gack, M. U. Dephosphorylation of the RNA Sensors RIG-I and MDA5 by the Phosphatase PP1 Is Essential for Innate Immune Signaling. Immunity 2013, 38 (3), 437–449. https://doi.org/10.1016/j.immuni.2012.11.018.
(5) Yu, Q.; Qu, K.; Modis, Y. Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis. Molecular Cell 2018, 72 (6), 999-1012.e6. https://doi.org/10.1016/j.molcel.2018.10.012. (PDB: 6G19)
(6) Wu, B.; Peisley, A.; Richards, C.; Yao, H.; Zeng, X.; Lin, C.; Chu, F.; Walz, T.; Hur, S. Structural Basis for dsRNA Recognition, Filament Formation, and Antiviral Signal Activation by MDA5. Cell 2013, 152 (1), 276–289. https://doi.org/10.1016/j.cell.2012.11.048.
(7) Xu, H.; He, X.; Zheng, H.; Huang, L. J.; Hou, F.; Yu, Z.; de la Cruz, M. J.; Borkowski, B.; Zhang, X.; Chen, Z. J.; Jiang, Q.-X. Structural Basis for the Prion-like MAVS Filaments in Antiviral Innate Immunity. eLife 2014, 3, e01489. https://doi.org/10.7554/eLife.01489.
(8) Watts, J. M.; Dang, K. K.; Gorelick, R. J.; Leonard, C. W.; Bess Jr, J. W.; Swanstrom, R.; Burch, C. L.; Weeks, K. M. Architecture and Secondary Structure of an Entire HIV-1 RNA Genome. Nature 2009, 460 (7256), 711–716. https://doi.org/10.1038/nature08237.
(9) Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; Žídek, A.; Green, T.; Tunyasuvunakool, K.; Petersen, S.; Jumper, J.; Clancy, E.; Green, R.; Vora, A.; Lutfi, M.; Figurnov, M.; Cowie, A.; Hobbs, N.; Kohli, P.; Kleywegt, G.; Birney, E.; Hassabis, D.; Velankar, S. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Research 2022, 50 (D1), D439–D444. https://doi.org/10.1093/nar/gkab1061.
(10) Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; Bridgland, A.; Meyer, C.; Kohl, S. A. A.; Ballard, A. J.; Cowie, A.; Romera-Paredes, B.; Nikolov, S.; Jain, R.; Adler, J.; Back, T.; Petersen, S.; Reiman, D.; Clancy, E.; Zielinski, M.; Steinegger, M.; Pacholska, M.; Berghammer, T.; Bodenstein, S.; Silver, D.; Vinyals, O.; Senior, A. W.; Kavukcuoglu, K.; Kohli, P.; Hassabis, D. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596 (7873), 583–589. https://doi.org/10.1038/s41586-021-03819-2.
